RNA structure and folding
study RNA folding using computer simulations
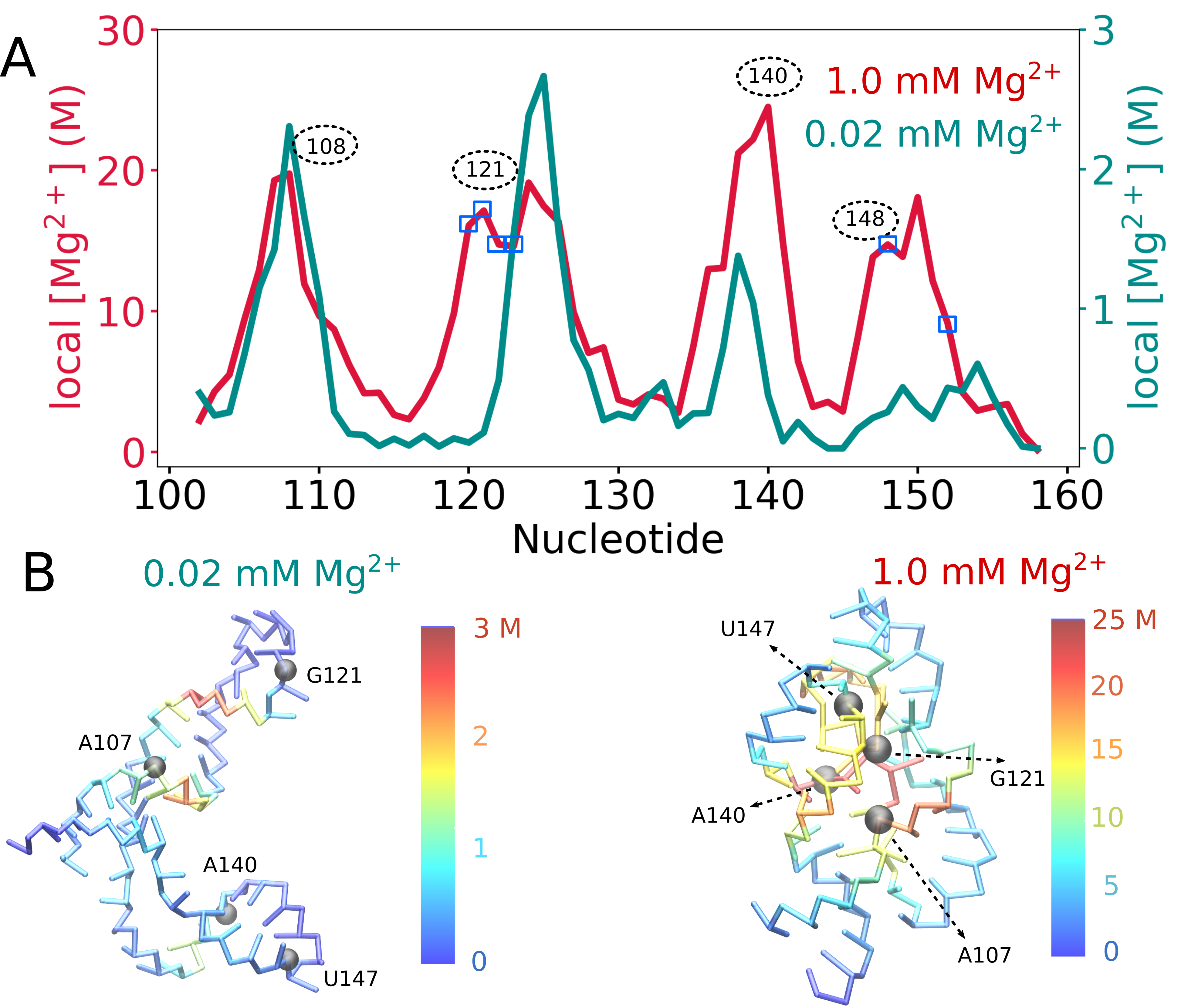
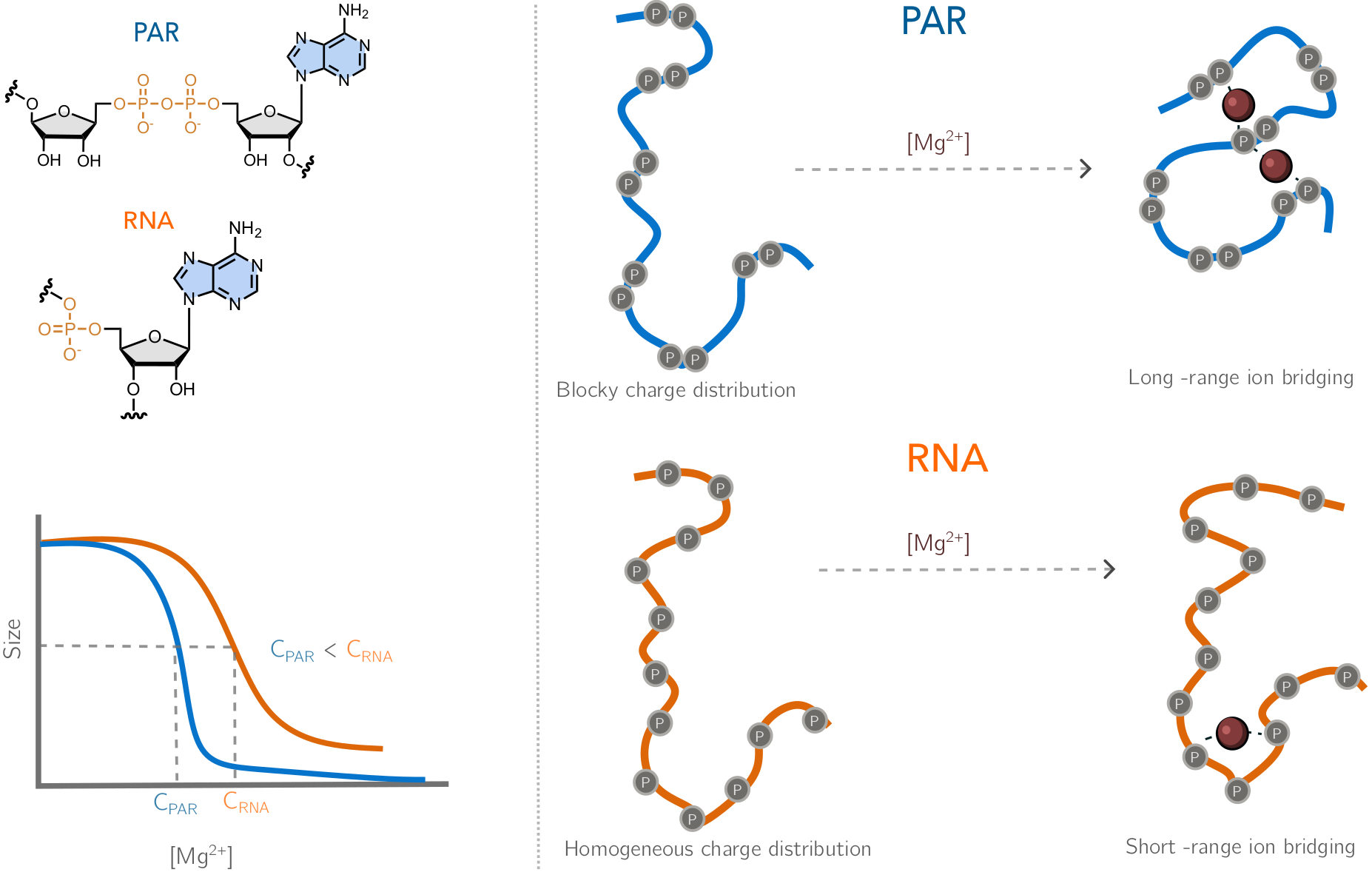
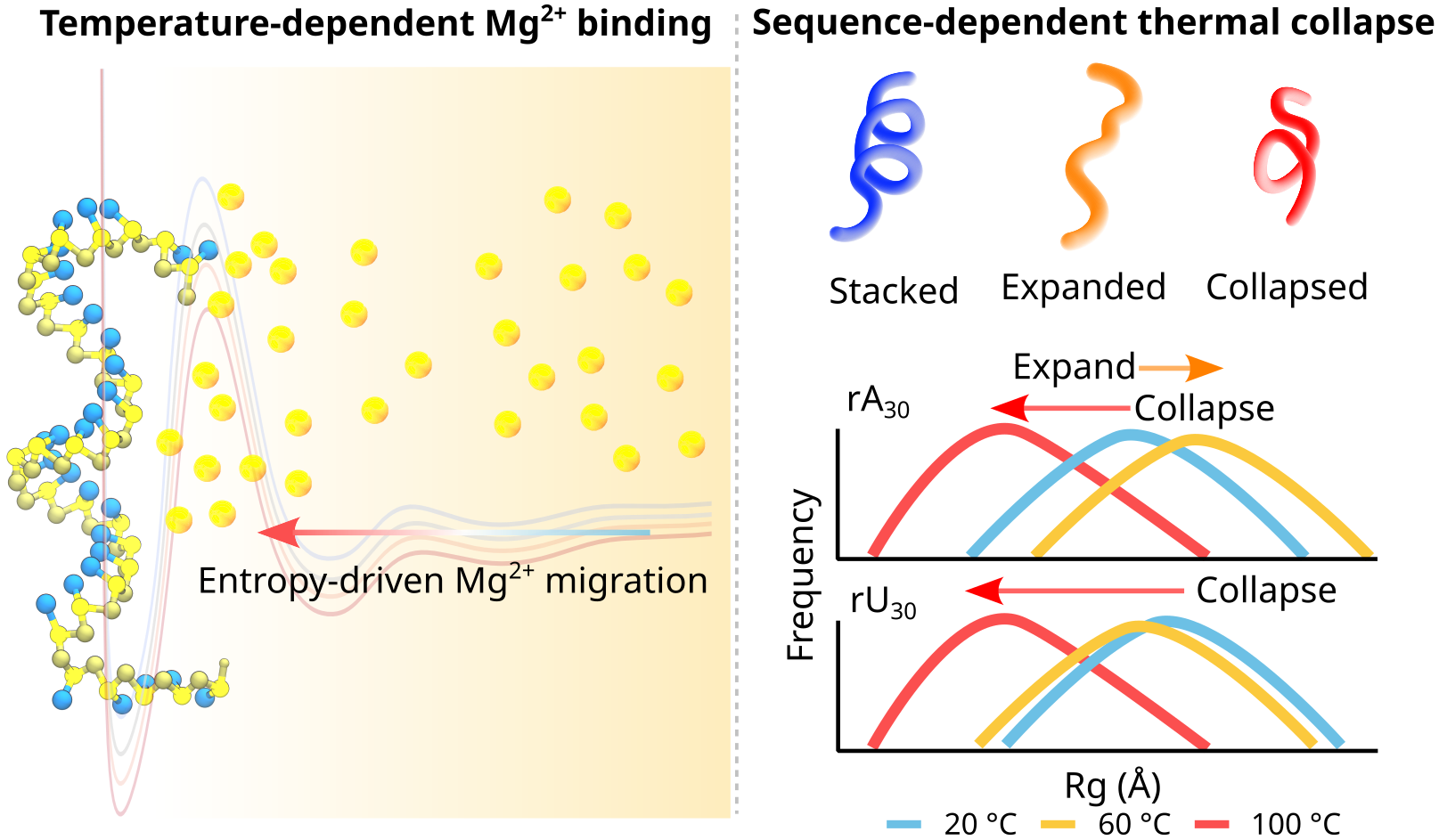
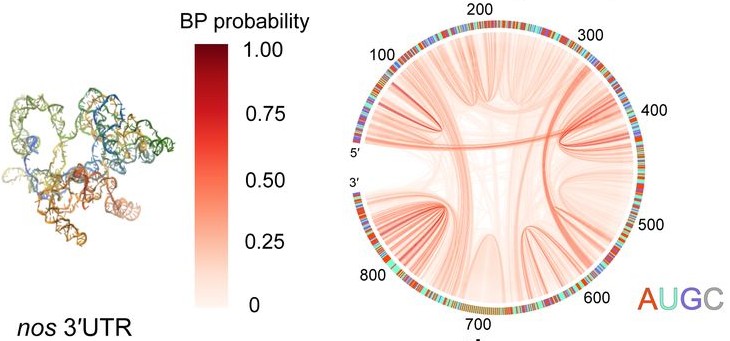
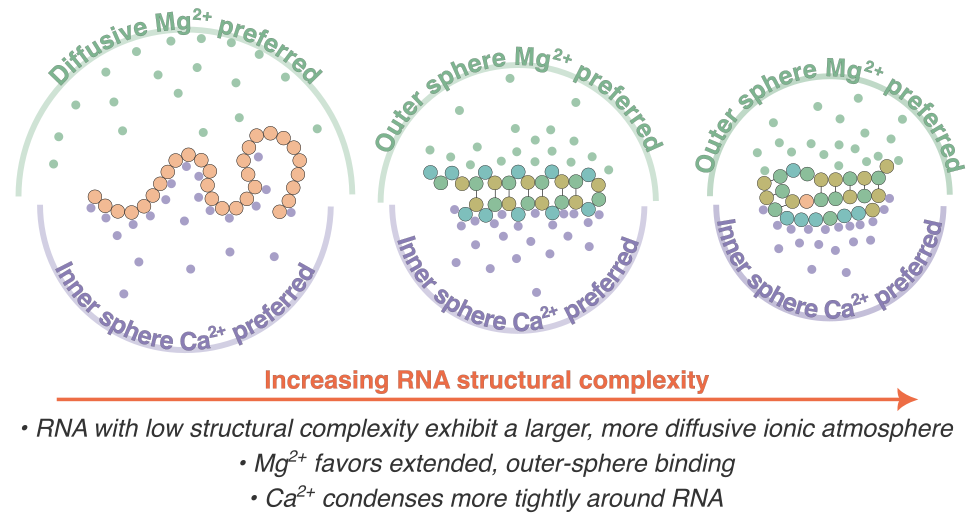
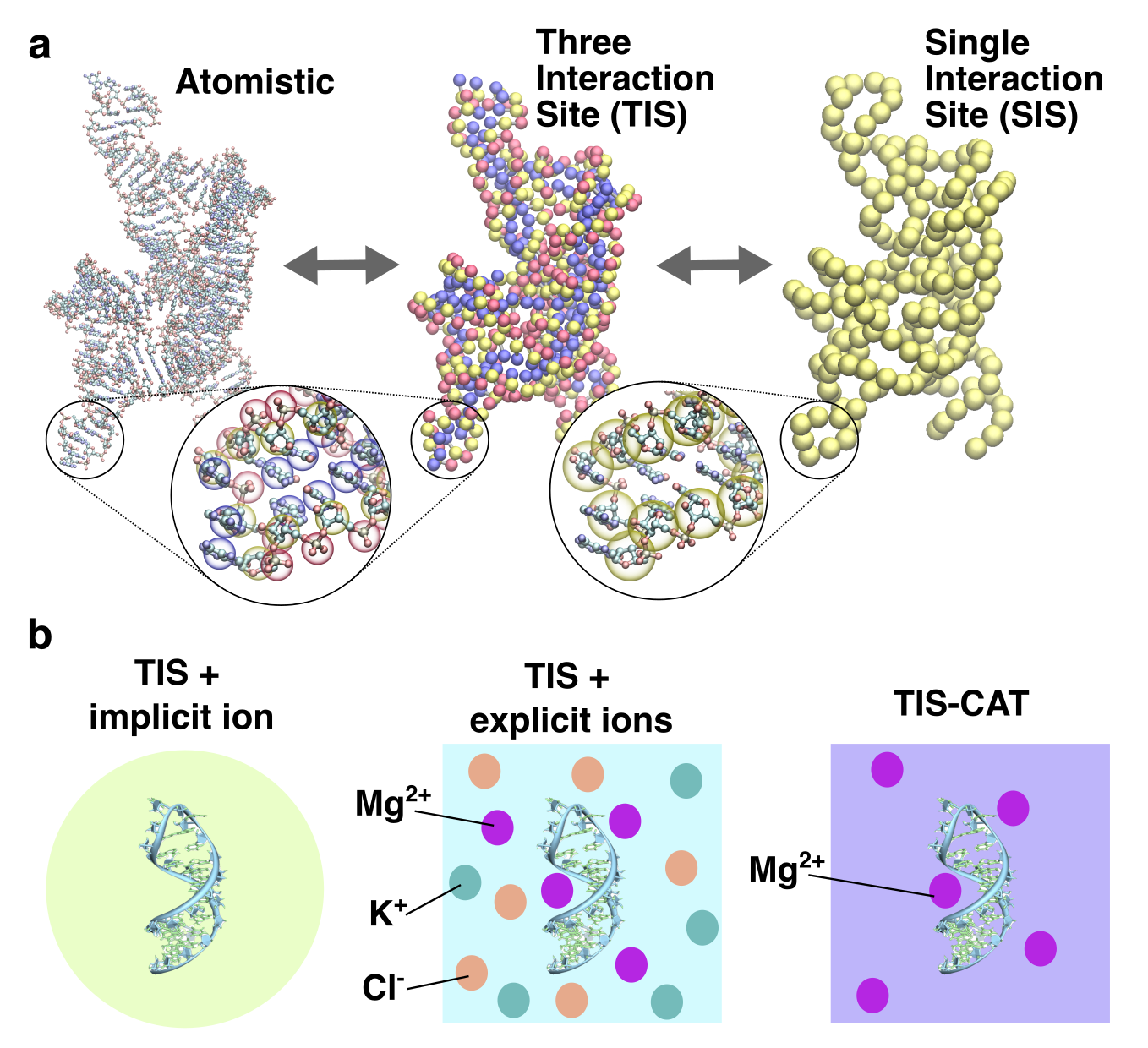
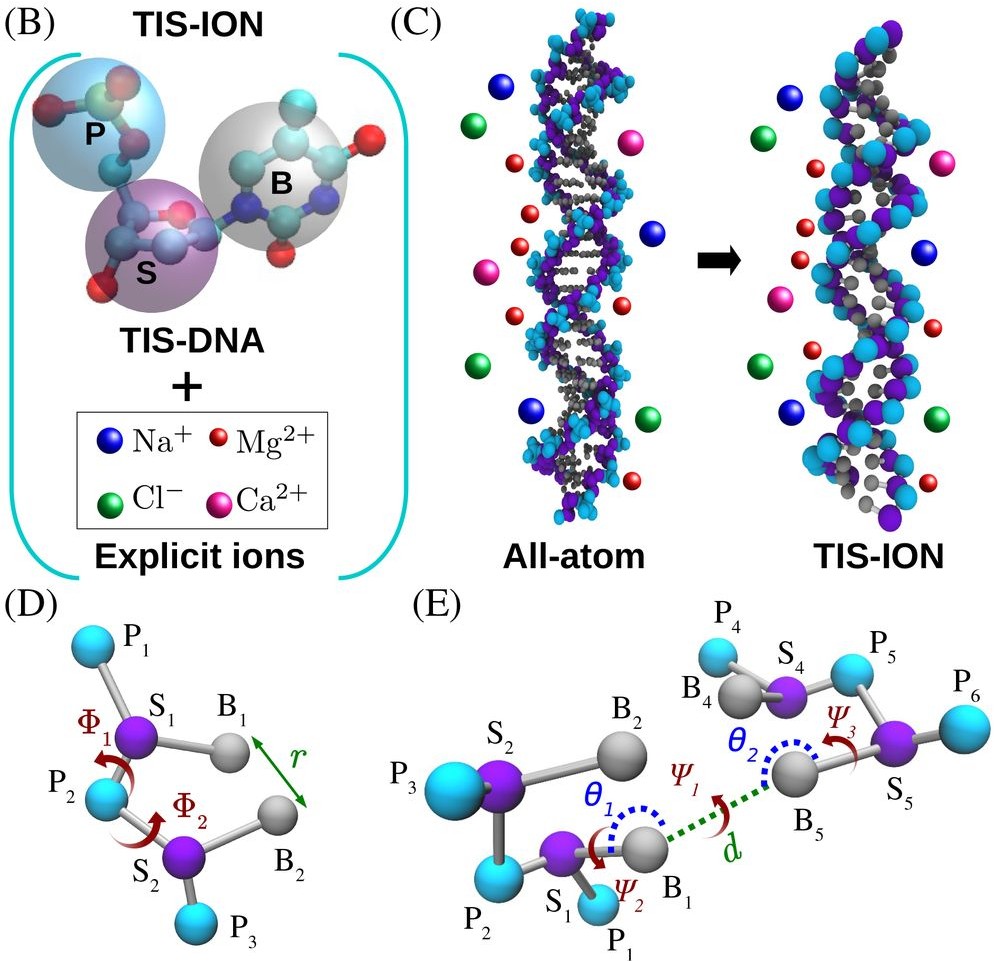
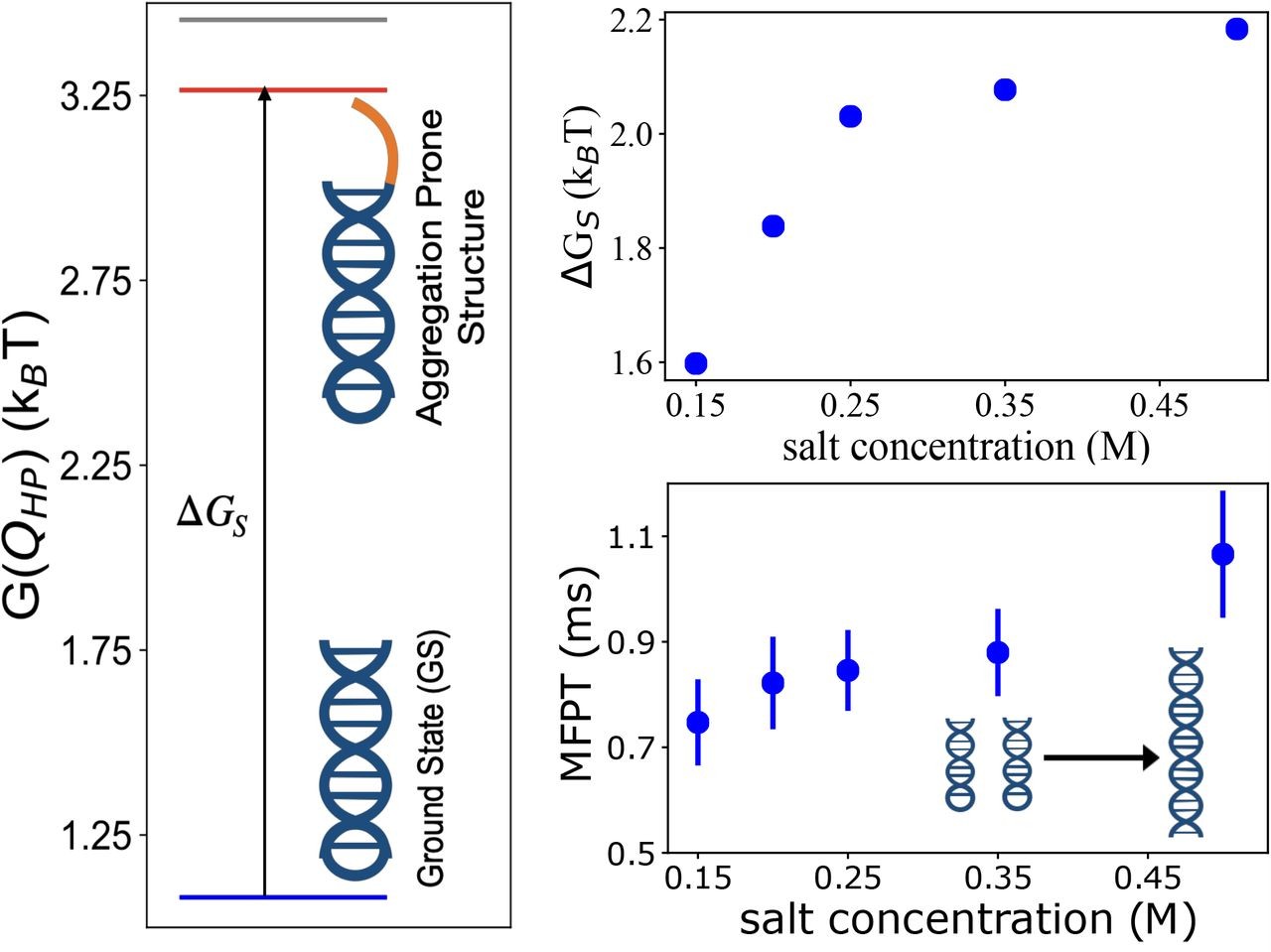
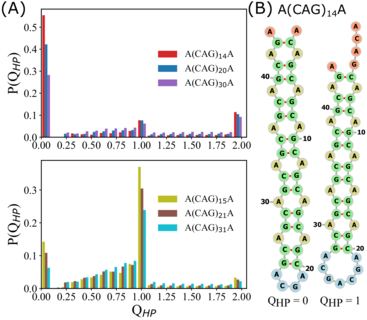
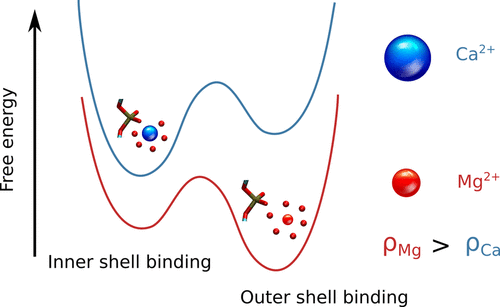
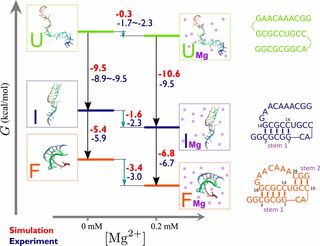
RNA–a highly charged biomolecule–can fold upon itself to form diverse secondary and tertiary structures to perform a vast array of cellular functions. Folding of RNA requires counterions, especially Mg2+, to reach a competently active state. Simulations of RNA folding in the presence of ions have been, and still are, demanding due to the long timescale and a large number of particles in the system. Using liquid state theory, we devised a method to treat monovalent ions implicitly, while retaining explicit divalent ions in the RNA folding simulations. This method drastically speeds up the simulations while maintaining their accuracy, allowing us to perform simulations to study the folding of larger and more biologically relevant RNAs. Additionally, the accurate treatment of ion-RNA interactions reveals the importance of Mg2+ in bridging specific RNA tertiary structures. Such molecularly detailed knowledge of how ions facilitate RNA folding deepens our understanding of the process, which could potentially be helpful in future RNA design and therapeutics. Other groups have used our method to investigate the folding of other RNAs, such as riboswitches.