BioSim Lab @ UB
Computational Modeling + Theory + Data Science
Welcome to the BioSim lab at the Department of Chemistry - University at Buffalo! Our team of researchers, led by Hung Nguyen, Ph.D., specializes in developing and applying computational methods & tools to study biological systems at the molecular level. By combining expertise in chemistry, biophysics, polymer physics & molecular biology, we aim to uncover the fundamental principles underlying the behavior of biomolecules, their interactions and functions. Our ultimate goal is to advance our understanding of biological processes and contribute to the development of new therapeutic approaches.
To learn more, check out our research and publications. We’re expanding our lab!!

contact us
| nguyenh [at] buffalo.edu | |
| +1 716 645 4166 | |
| 471 Natural Sciences Complex, Buffalo NY, 14260 |
news
| Feb, 2026 | Our group was going full force for the annual BPS meeting at San Francisco CA. Three members of the lab (Peter, Lipika and Dibya) gave posters for their work. Lipika got the travel award for her poster about poly(ADP-ribose) structure and phase behavior, which hopefully appears in print soon! |
|---|---|
| Feb, 2026 | Delighted to contribute to an editorial on J. Med. Chem. about targeting RNA using small molecules. Thanks D. Heppner and others for the invitation! |
| Jan, 2026 | This month we welcome Bola to the lab! |
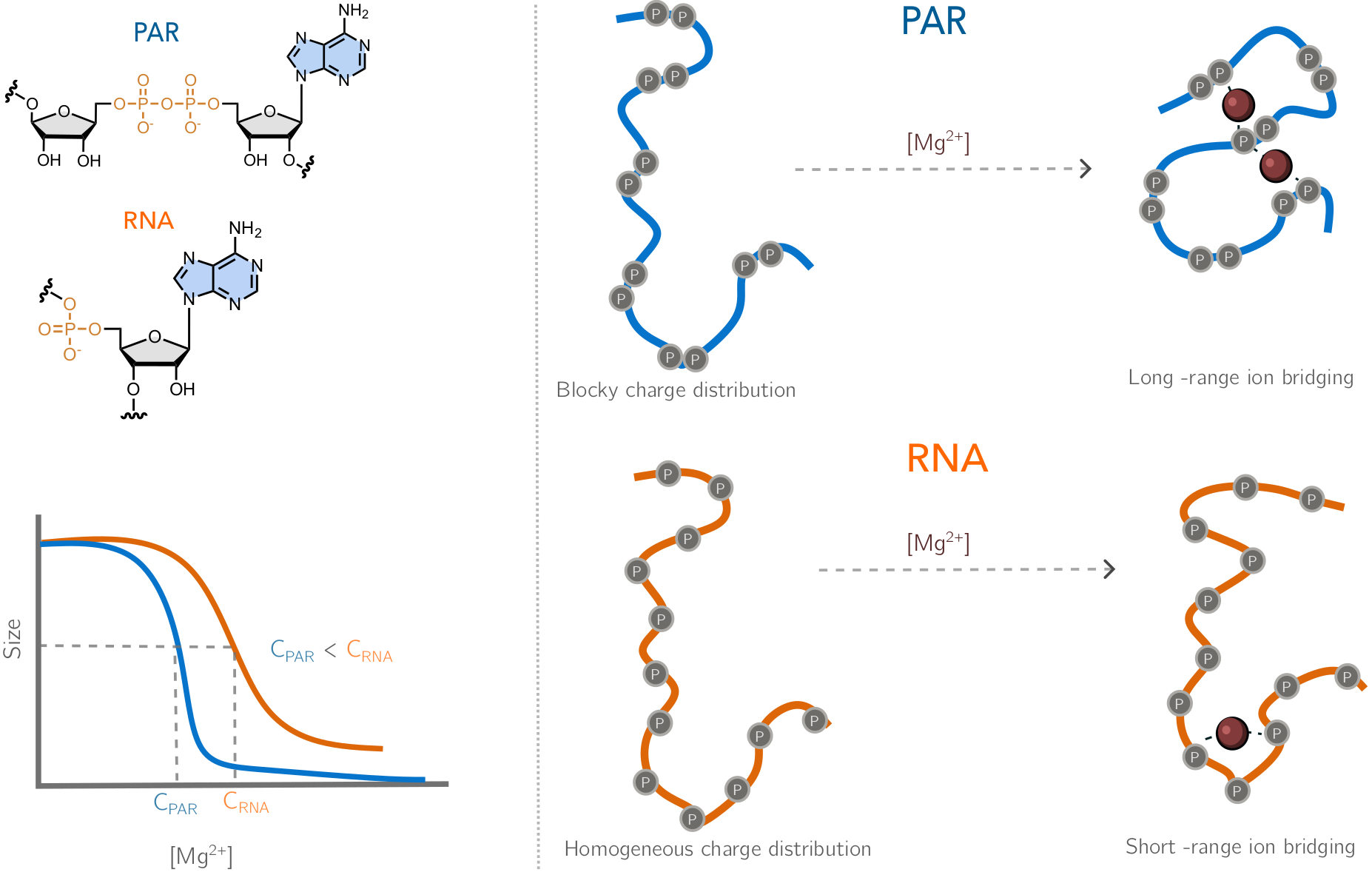
| Dec, 2025 | Another month, another exciting finding! Lipika’s work about modeling of poly(ADP-ribose) (PAR) is on biorXiv. We introduce a novel coarse-grained model for PAR and show that despite sharing the chemical composition with RNA, PAR is a completely different beast! Exciting new direction … |
| Oct, 2025 | Three members of the lab (Peter, Lipika and Dibya) gave posters at Rustbelt RNA Meeting (Ohio) for their work. Lipika got the travel award for her poster about poly(ADP-ribose)! |
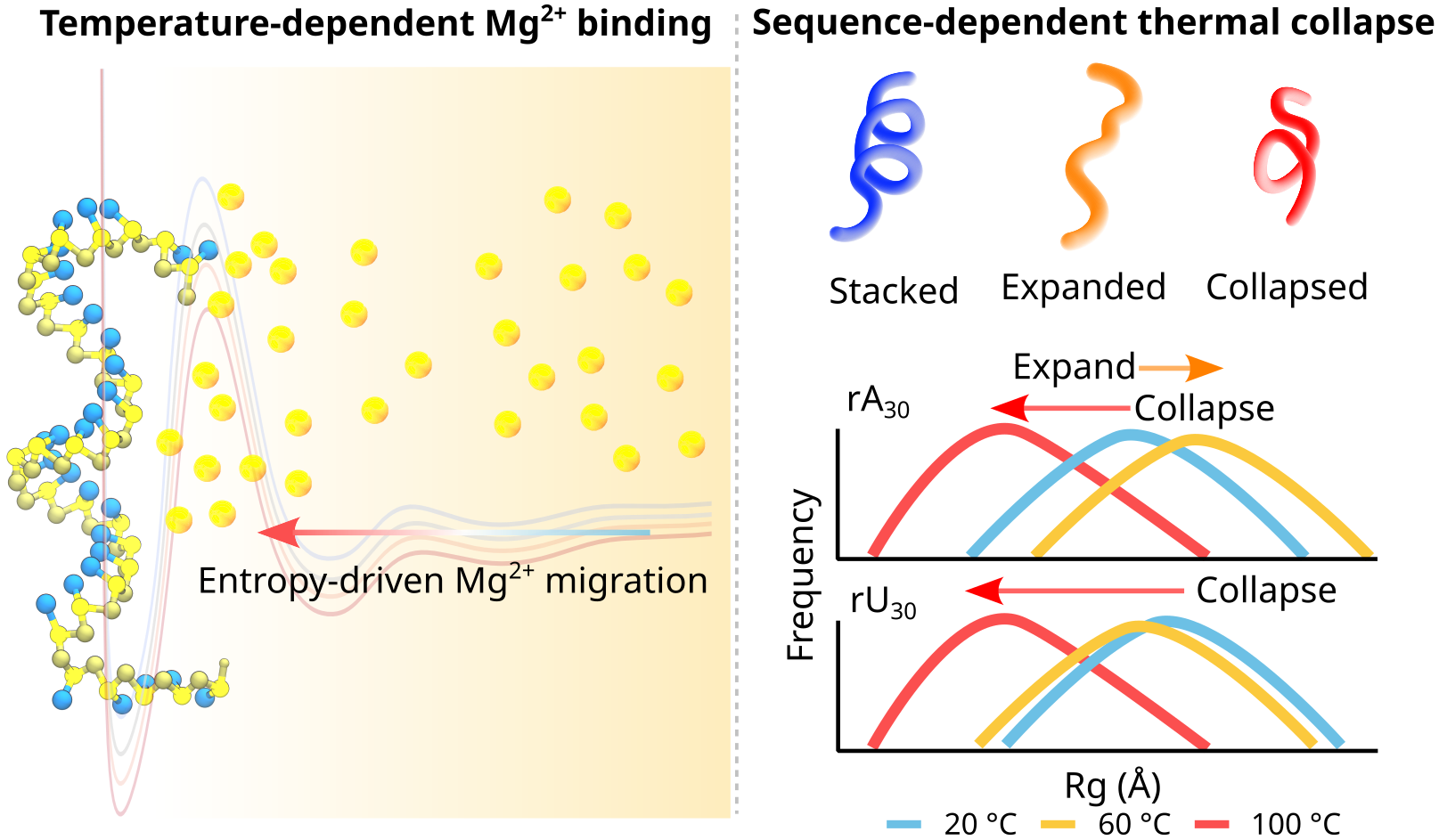
| Oct, 2025 | Super excited to send Peter’s work out biorXiv. We show that temperature and counterions jointly affect RNA conformational ensemble, and more excitingly, high temperature leads to the collapse of RNA ion atmosphere. Watch this space! |
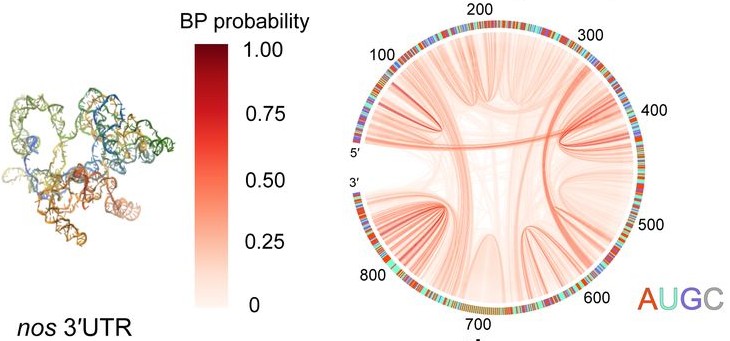
| Sep, 2025 | After a long time, this work has been published on Nat Comm. With collaborators we show that RNA structure impedes intermolecular base pairing in germ granule. |
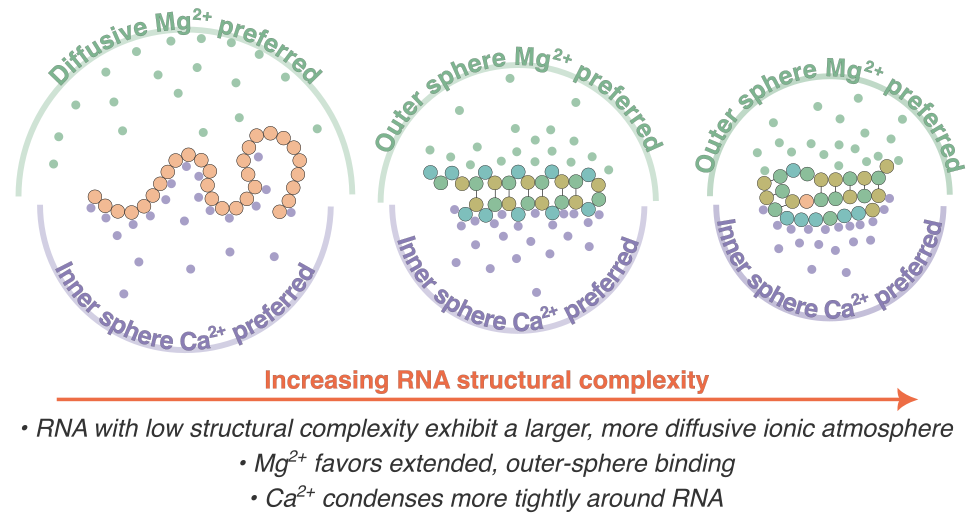
| Aug, 2025 | Excited to share our lab first published paper spearheaded by Hiranmay and Peter on J Phys Chem Lett. We demonstrated that the ion clouds of unstructured RNAs extend further in the solution than those of folded domains, suggesting a role for disordered RNA to be a recruiter using long-ranged interactions. |
| May, 2025 | Delighted to send our first lab manuscript out to bioRxiv! |
| Mar, 2025 | After a long wait, our manuscript about IDP modeling has been accepted to Protein Science! |
| Mar, 2025 | Hung was invited to speak at the ‘Multi-scale Modeling of Biomolecular Condensates’ symposium during the ACS Spring ‘25 Meeting in San Diego, CA. |
| Feb, 2025 | Peter presented two posters at the Biophysical Society Meeting in LA, showcasing his research on RNA structural ensembles and their phase behavior |
| Jan, 2025 | Ecstatic to welcome THREE new members joining the lab |
| Dec, 2024 | Check out our two new preprints on RNA directing physical properties and functional outputs of biomolecular condensates BioRxiv and a review about RNA modeling arXiv. |
| Oct, 2024 | Peter and Hiranmay gave posters at NY RNA Symposium for their work about modeling RNA structural ensemble and phase separation propensity. Peter got an outstanding award for his poster! |
| Aug, 2024 | Hung gave a talk at ACS ‘24 Fall, Denver CO. It was nice to catch up with old friends & colleagues, and meet many new potential collaborators. |
| Aug, 2024 | Dibya joins us as the second postdoc in the lab |
| Jun, 2024 | Check out our new preprint on BioRxiv, which is a collaborative effort with the Trcek’s and other labs. We showed that mRNA retains most of its structure inside germ granule, and that RNA-RNA homotypic interactions are limited, and are mostly formed by scattered intermolecular base pairing. |
| Apr, 2024 | The first undergrad joins the lab, woohoo. |
| Apr, 2024 | Two manuscripts have been accepted to J. Phys. Chem. Lett. and J. Chem. Theory Comput.. Congratulations! |
| Dec, 2023 | Delighted to welcome TWO new members joining the lab |
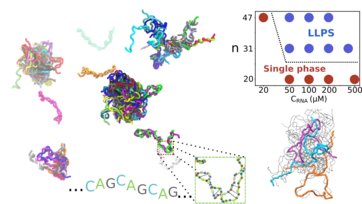
| Dec, 2023 | Check out our new preprint on BioRxiv about salt dependence of the self-association propensity of CAG repeats! |
| Oct, 2023 | Super excited to welcome Peter, the very first grad student, joining the lab! |
| Oct, 2023 | Check out the preprint from Thirumalai’s lab (with our small contributions) about DNA modeling in the presence of ions bioRxiv! |
| Aug, 2023 | Hung got invited to give a talk at the Genome, Environment and Microbiome (GEM) Community of Excellence Work-in-Progress in the Jacobs School of Medicine and Biomedical Sciences, UB. |
| Aug, 2023 | The BioSim lab officially starts at UB! |
| Jun, 2023 | Our recent work has been accepted and now appears on PNAS! We establish a link between structures & thermodynamics of monomeric RNAs and their propensity to undergo phase separation & aggregation. |
| May, 2023 | Hung gives talk at multiple universities in Vietnam |
| Apr, 2023 | Check out our work of IDP on bioRxiv! Also, try out the code to run simulations using OpenMM with GPU acceleration. |
| Mar, 2023 | Hung has been offered a tenure-track Assistant Professor position in the Department of Chemistry - UB! Excited to start the new lab Fall ‘23! |