interactions between solvent and biomolecules
extract solvent distribution from SAXS and computer simulations
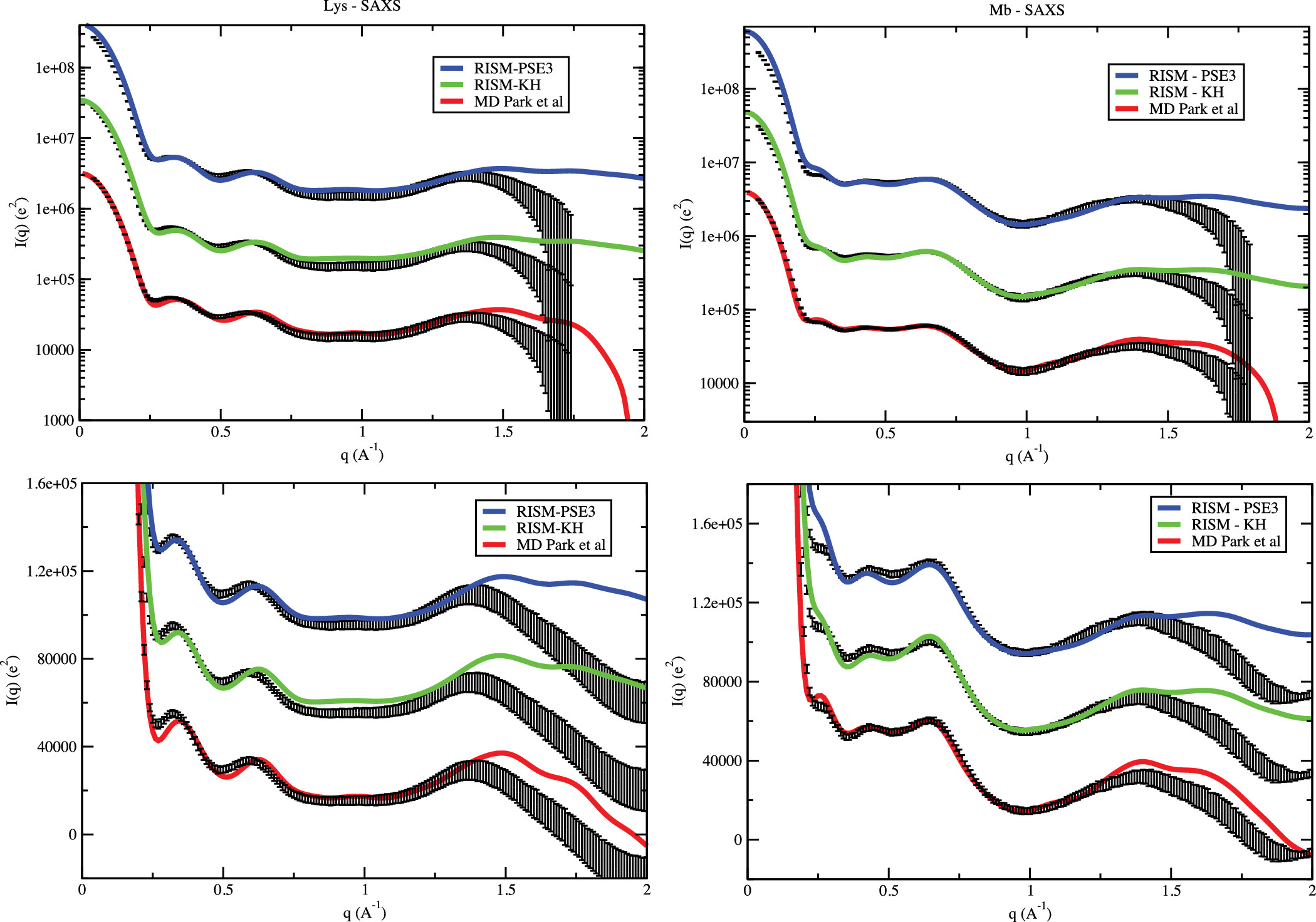
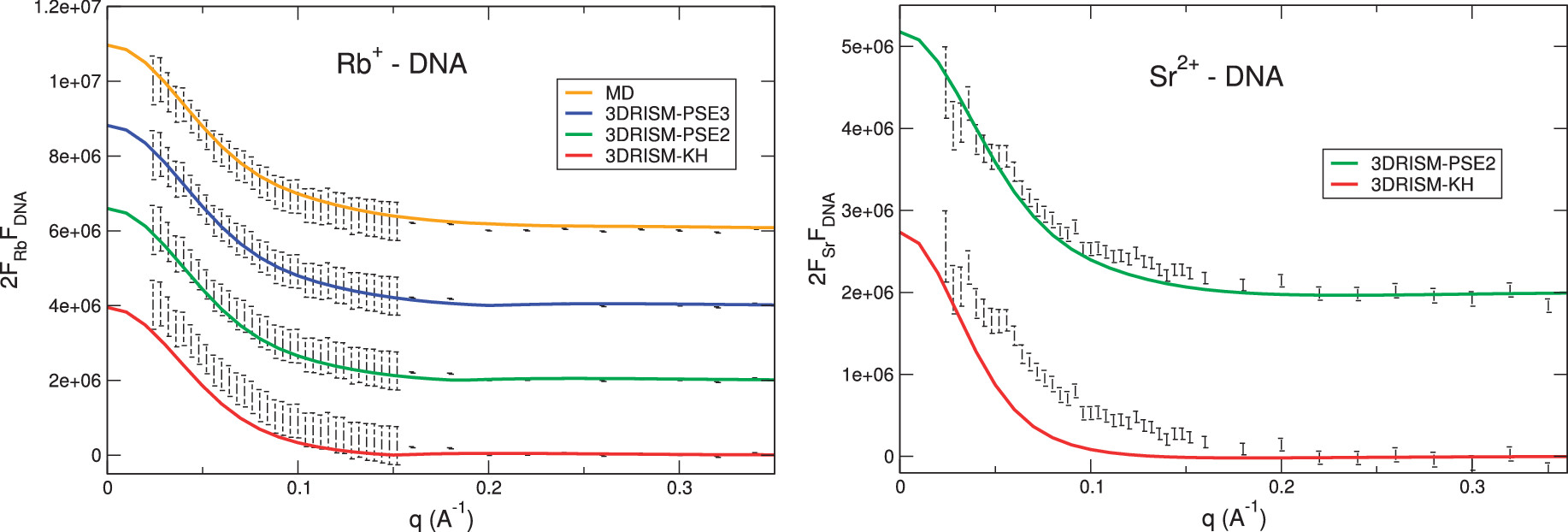
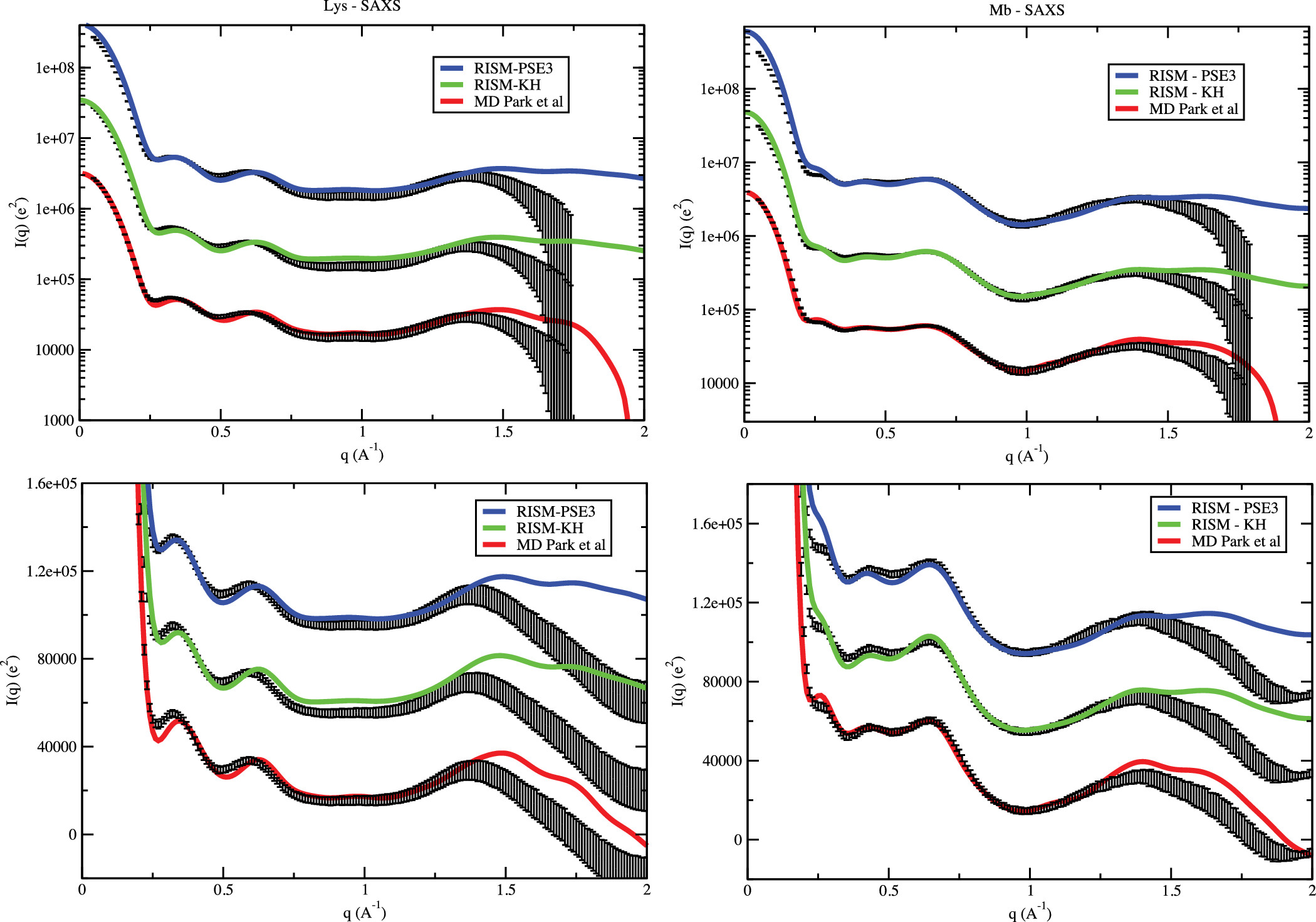
We focused on understanding the interactions between solvent and proteins & nucleic acids, and characterizing the spatial distribution of solvent around biomolecules. It has been long known that both water and counterions, which carry the opposite charges to the biomolecule, accumulate at higher concentrations in the vicinity of the biomolecule’s surface. However, exactly how the distribution takes shape is still elusive. Using computer simulation and liquid state theory, we demonstrated that in addition to the number of ions usually measured in solution X-ray scattering or "ion counting" experiments, the 3-dimensional spatial distribution of solvent could be obtained faithfully. The biomolecule, therefore, could be thought to carry the hydrated and ionic atmosphere around as a molecular "fingerprint". This fingerprint extends quite far away from the biomolecule surface and therefore could facilitate long-ranged molecular recognition. In addition, we have proposed a new method to extract some aspects of the solvent spatial distribution directly from X-ray scattering profiles, expanding the information obtainable from the experiment.